Automating research data flows
Table of Contents
For most of my career I've relied on cronjobs to run my scheduled automations. Whenever I need to add robustness I rely on heartbeat alerts and if I need version control or continuous integration I use GitHub self-hosted runners to automatically deeply updates on commit. However, I joined a project a few years ago where I created custom automations that I wanted to allow the non-engineers in my team to trigger manually or with custom arguments. Additionally, these automations were hosted across multiple different geographical regions so building a simple web GUI would've been insufficient.
Building on cronjobs
In usual research projects we typically have a single server which we use for data management and also scheduling cronjobs. The server is monitored using our center's prometheus/grafana stack which generates an alert if there is an outage. I would make the cronjob to the local server's /var/log directory and if a daily job didn't run for a few days I'd ssh in to the server and check the logs. This was a very reliable arrangement.
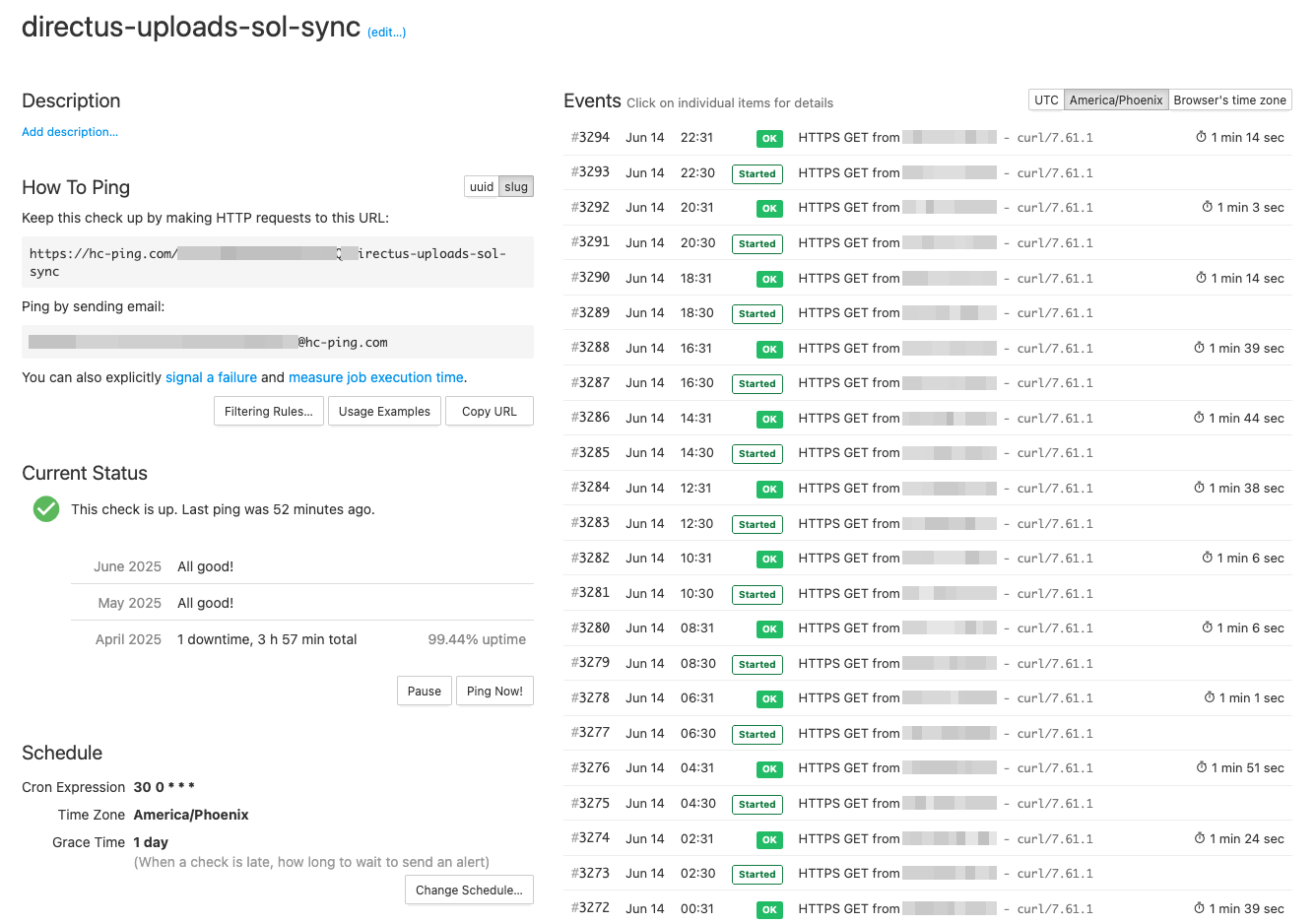
Eventually, we had automations that interacted with other unreliable remote systems so the cronjobs would fail more frequently with different errors. To better monitor these jobs I rely on an amazing tool Healthchecks.io with a generous free tier. Healthchecks is a heartbeat style monitoring service where you can add generate unique ping URLs for each job.
#!/bin/bash
curl -fsS -m 10 --retry 5 -o /dev/null https://hc-ping.com/<unique_job_url>/start
echo "So sleepy"
sleep 10
echo "Fully refreshed!"
curl -fsS -m 10 --retry 5 -o /dev/null https://hc-ping.com/<unique_job_url>/$?
Heartbeat monitoring works on the principle of "expected check-ins." If your job doesn't ping the monitoring service within the expected timeframe, you get alerted. It's simple and effective.
It also supports the ability to send a start ping and an end ping with the optional ability to record exit code. In addition to failed runs, this allows us to track the run time of our jobs. It also supports receiving 'pings' over email which I use for our automations that run on alteryx gallery.
Web based data applications
In addition to the scheduled automations, we also have a category of jobs that have to be triggered manually and they produce an output for the user to download. For example, in a hospital we want to create wristbands in advance for people who checked in at a remote field site or in a lab we want to generate new unique identifiers for our vials from a biosample database. For these sorts of tasks we've relied on creating custom Shiny apps.
Shiny apps is a web framework created by Posit intended for developing data related interactive web applications. The framework used to only support R, though in 2022 they've also added support for python. These apps can be hosted on a managed cloud provided by posit or self-hosted on an open source Shiny Server. Since data scientists in our labs primarily use R or python, the Shiny apps fit our use case perfectly.
In our projects, either I will take someone's existing R code and turn it into a shiny app or the data scientist would themselves develop the shiny app. In either case, I was responsible for deploying them and my go to method is to use github for storing the application code and using a self-hosted github runner for auto deployment on every commit.
name: Deploy Shiny App
on: [push]
jobs:
deploy:
runs-on: self-hosted
steps:
- name: Checkout Repository
uses: actions/checkout@v2
- name: Write Commit Hash
run: echo $GITHUB_SHA > ${GITHUB_WORKSPACE}/commit_hash.txt
- name: Deploy Shiny App
run: |
rm -rf /srv/shiny-server/* && rsync -av --omit-dir-times --no-perms --no-group --delete --exclude '.git' --exclude '.github' ${GITHUB_WORKSPACE}/ /srv/shiny-server/
The
Write Commit Hashpart writes the hash to a text file which I display on the shiny app itself. Since most updates only changed the backend code without touching the frontend, this hash was a way to let the users know the version of the application they were interacting with. We use cloudflare zero-trust tunnels to expose our internal applications to authenticated users. Cloudflare has aggressive caching so viewing the hash also assisted with diagnosing cache related issues.
This was perfect for me since I did not have to be in the loop each time the data scientists would need to update the shiny apps.
Wanting the best of both worlds
Eventually, we ran into a situation where we wanted to reliably run automations across multiple geographical regions and occasionally with custom arguments. My ideal requirements for the tool were:
- No vendor lock-in: We should be able to run our cronjobs as is without making any changes.
- Centrally managed: Ability to run jobs across multiple servers from a single instance.
- Logging: Maintain an audit log for each run along with the stdout of the scripts.
- Web GUI: To allow non-programmers in the group to run the jobs with custom arguments.
I explored dagster, prefect, luigi, windmill and apache airflow and the major issue with all of the tools were that they (for good reason) did not support bash which meant additional effort in our part to rewrite the jobs in python. The tool that I did come close to adopting was apache airflow just because of its popularity however after a little experimentation I could tell that it'd require multiple hours of effort to properly learn apache airflow and then implement for our jobs.
I essentially wanted a smarter cronjob manager and the tool that fit all of my requirements was windmill. The initial migration to windmill was effortless. I copied my scripts verbatim to windmill and just added the schedules and et voila! I had all the logging that I wanted. Adding a GUI for allowing custom arguments was also straight forward for both bash and python scripts.
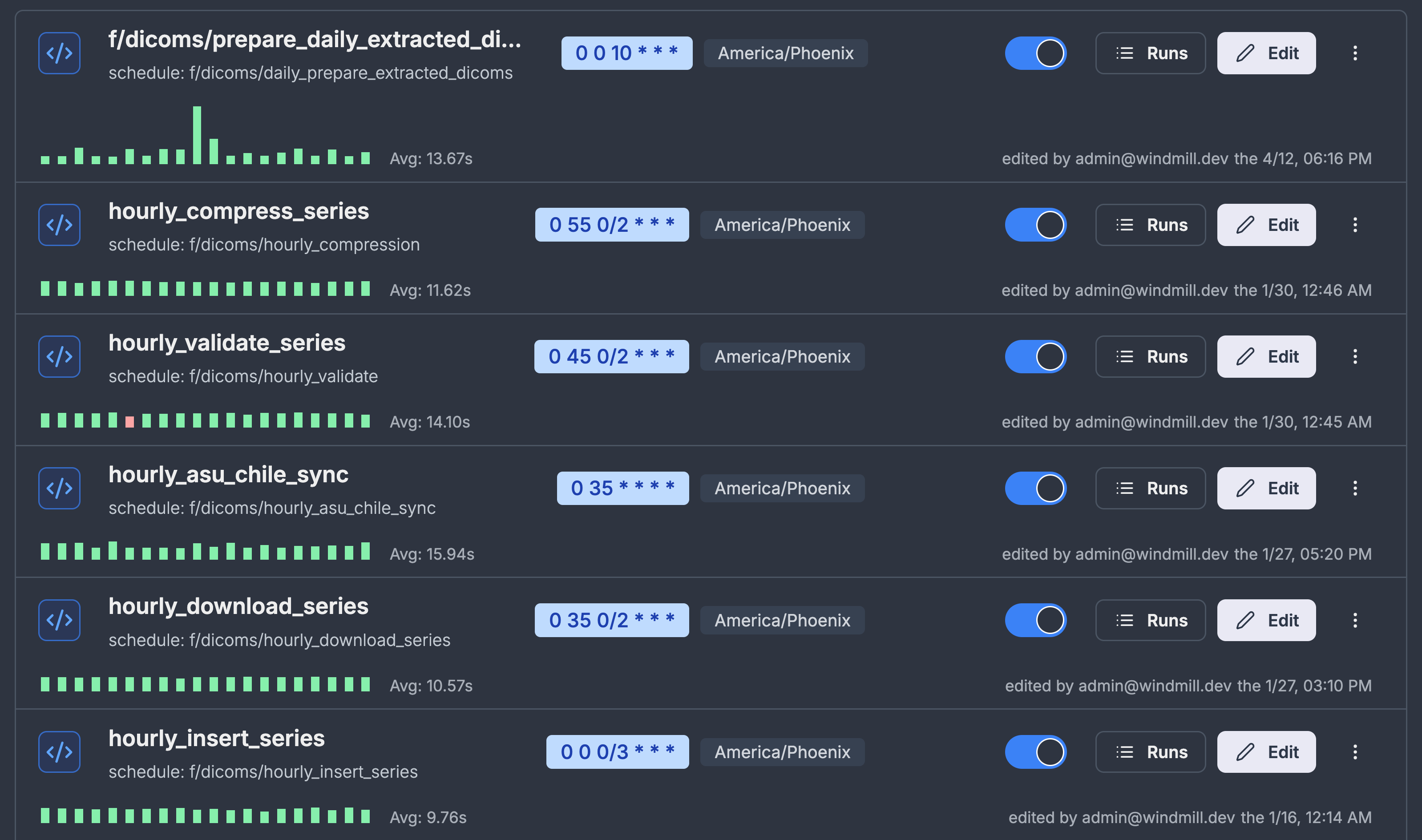
Another bonus feature within windmill was that I could store credentials to all my resources and directly use them within my scripts. Historically, this has been a pain point for me to either rely on .env file or an external file containing credentials just so we don't accidentally commit credentials to our git.
Windmill also has a bunch of other features like the ability to make custom web apps which we see great potential in to replace our shiny apps. With built-in oauth we also don't have to worry about rolling out our own authentication unlike the shiny apps. Overall, windmill checks all the boxes for us.
Use case: Windmill's role in THLHP
The Tsimane Health and Life History Project (THLHP) is a longitudinal biomedical study focused on understanding healthy aging in a non-industrialized population, specifically among the Tsimane indigenous group in Bolivia's lowlands. Since its inception in 2001, the project has collected extensive demographic, genetic, biochemical, and medical imaging data to investigate chronic diseases of aging, such as Alzheimer's and cardiovascular disease, resulting in a unique dataset spanning over two decades. As the project continues to grow and generate vast amounts of complex data, efficient management and analysis of this data have become increasingly important, which is where tools like windmill come into play.
BBC Documentary: The Tsimane Tribe
Learn about the Tsimane indigenous group and THLHP studying their health and aging patterns in Bolivia's lowlands.
Form submissions to data lake
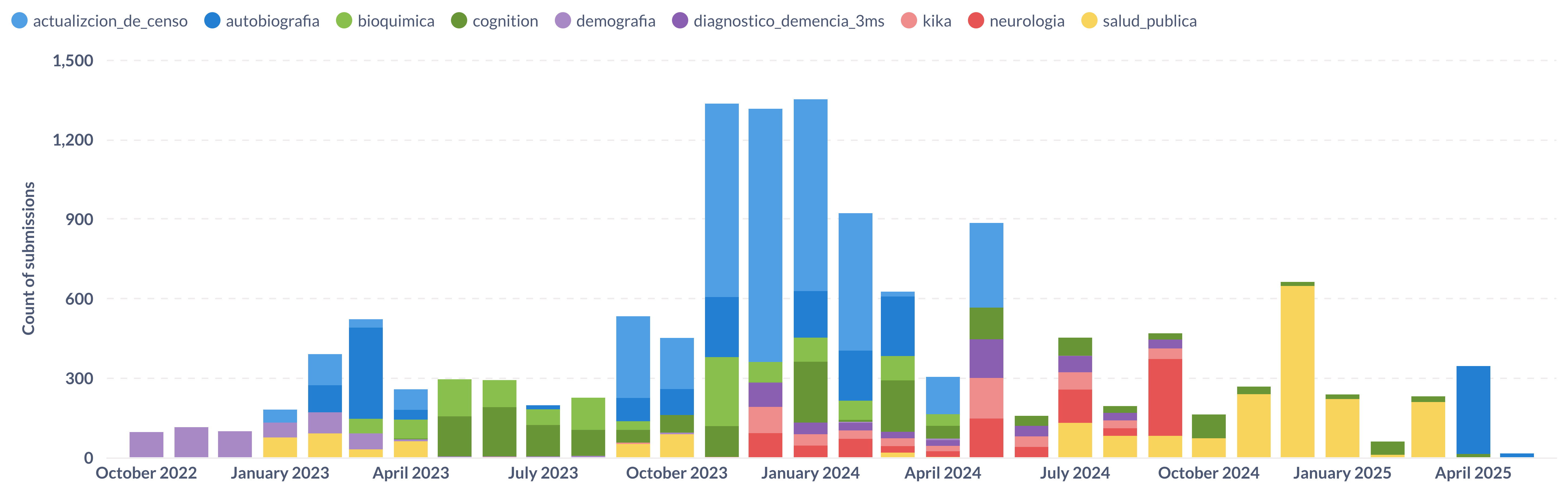
As part of effort to digitize data collection and move to a data lake model, the project recently switched from Microsoft Access to KoboToolbox. KoboToolbox provides us with the ability to collect data offline, push updates to questionnaires and have the forms available in multiple languages. KoboToolbox stores the form submissions in a mongodb database and provides access to it through their API.
To regularly import data from Kobo into our data lake we have a simple python script that takes in the kobo API endpoint as an argument and imports all submissions to our postgres database. We store a list of all our endpoints, database credentials and API tokens as windmill variables and then rely on windmill flows to iterate over each endpoint to populate our database. Each time we have a new form or endpoint, the only component we need to update is the windmill variable and the new submissions start coming in. Another benefit of using windmill is getting more granular logging over each run of the script with different arguments.
Radiology transfers and management
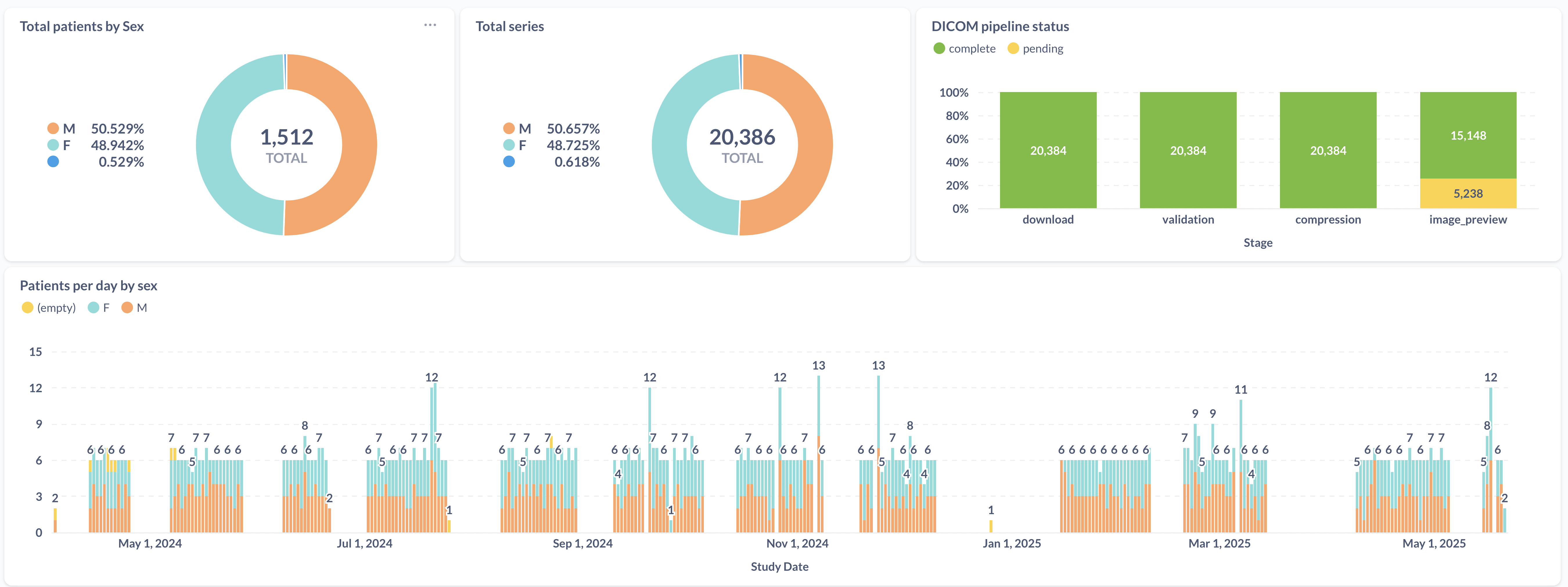
The project collaborates with a local hospital in Trinidad Bolivia where CT-scans are conducted. CT scans are copied over to the hospital's Picture Archiving and Communication System (PACS) as DICOMs and from there we copy the relevant research scans to the project's data repository. The data transfer pipeline consists of multiple jobs that run independently, they query, download, validate, extract metadata, compress, and transfer data. They interact with each other by using our postgres database as a job queue to check for pending tasks.
DICOM (Digital Imaging and Communications in Medicine) files are a standard format for medical imaging. A DICOM file in addition to containing the pixel information of the image in binary format also contains all the metadata relating to the machinery, the protocols, the patient and the hospital. Each scan can produce hundreds of .dcm files with each containing information about a single slice. Hence, managing DICOMs directly over the filesystem without a PACS is a complex task.
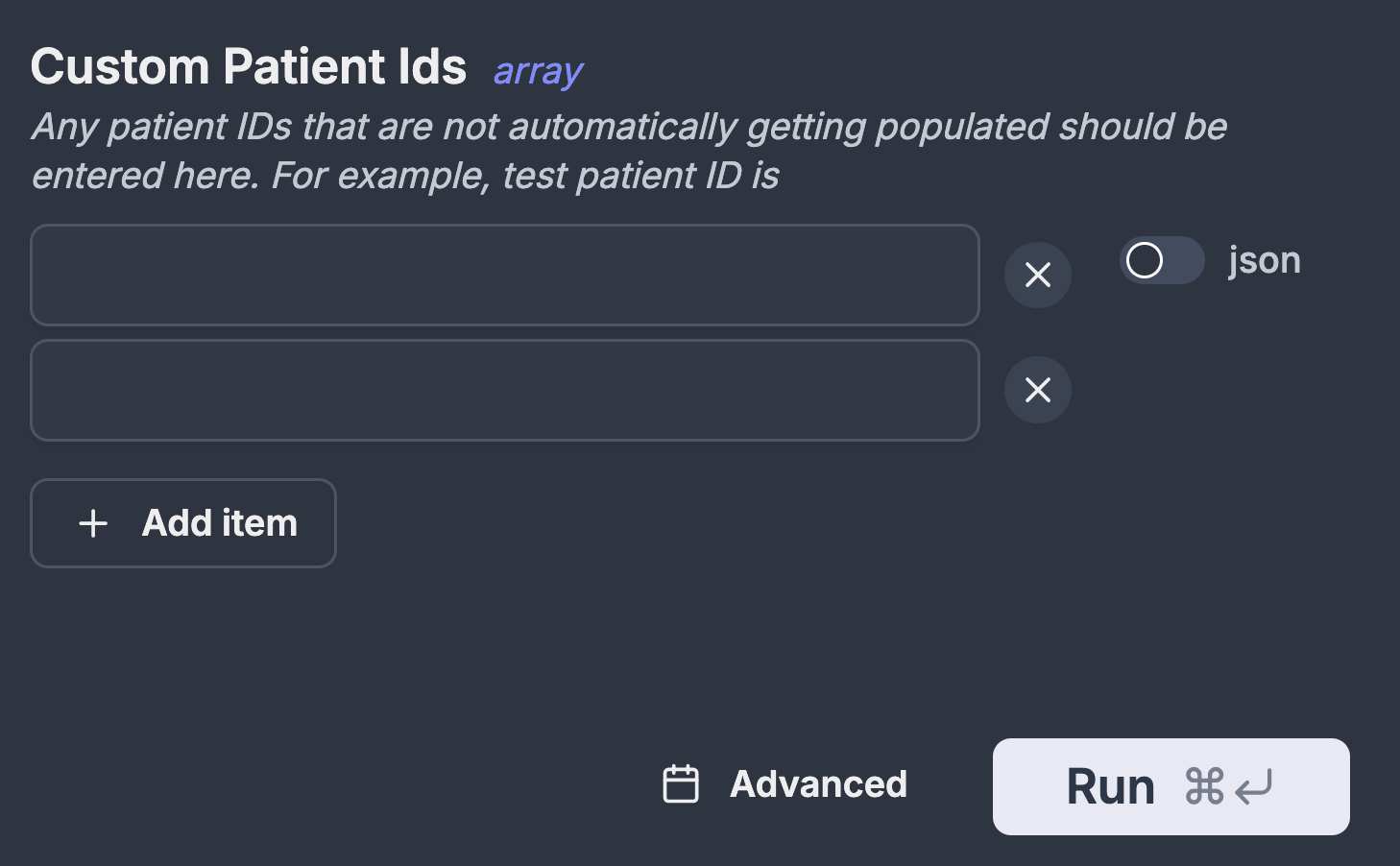
The first job that kicks everything off is the automation that checks the hospital's PACS for any new data from THLHP participants. The job identifies THLHP participants by using a regular expression. If a new patient is detected then their information is added to our central database which kicks off downstream jobs to download any data relating to them. Occasionally, there are circumstances when we want to download data for participant_ids that do not match our regex, for example when there is a typo or when the machine is being calibrated. For these circumstances the automation accepts custom custom_patient_ids as a list which can be populated through windmill GUI.
Since everything is coordinated over postgres, we have a nifty dashboard to track the progress of scans as they are download and made available in our data repository.
Data validation
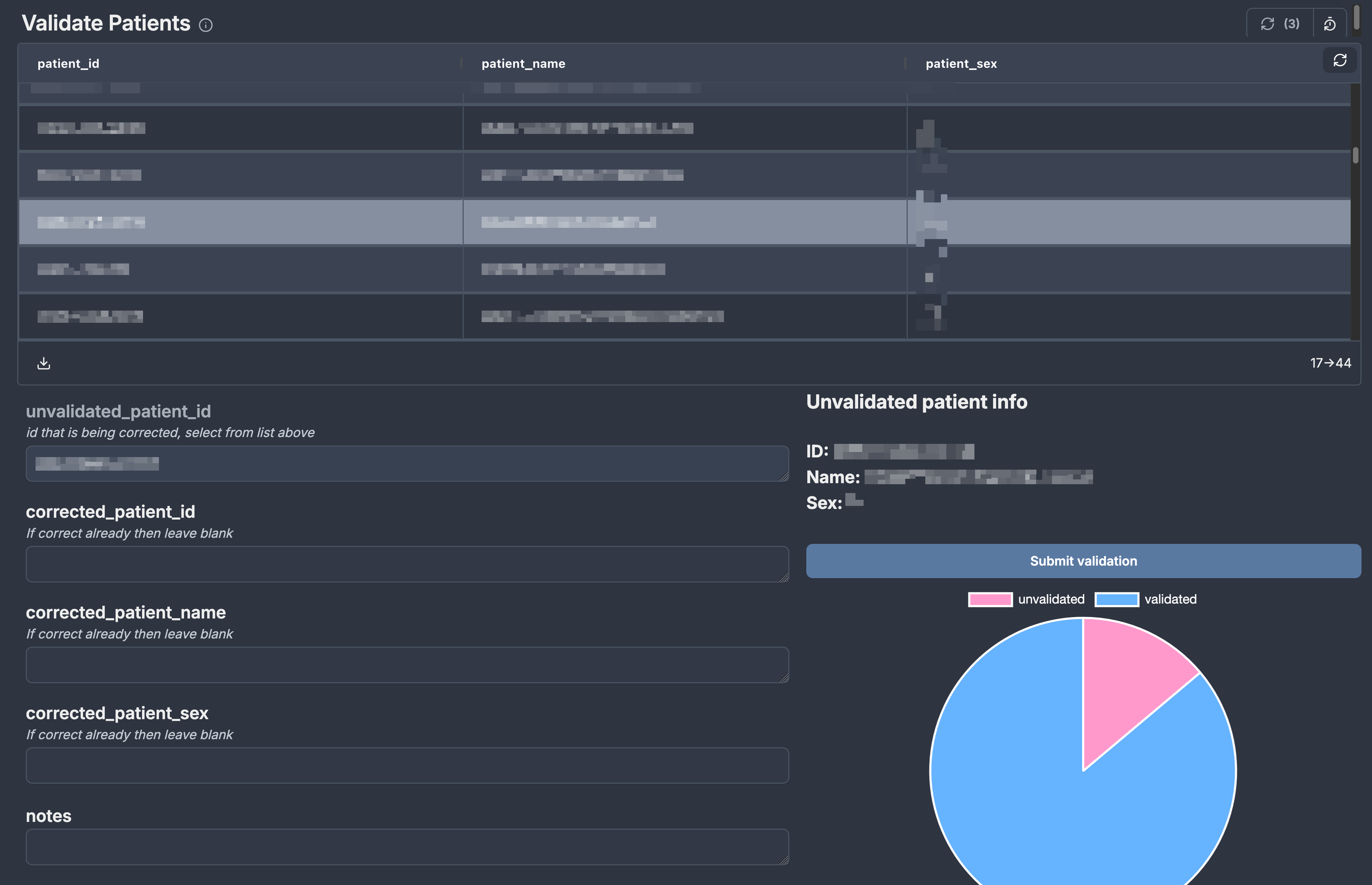
Automated scripts check the expected patient IDs against the IDs manually entered by hospital technicians to ensure there are no errors. If typos or misspellings occur, we created a mechanism where a second person could validate the entered data and submit any corrections without altering the source data. For this purpose we experimented with windmill to create a simple web app that queries all the unvalidated patients and allows anyone to submit a correction.
This solution worked out splendidly without us having to learn any javascript or other frontend languages. Another benefit of using windmill to submit a correction was automatically recording the name and email of the person submitting the correction since we use oauth to secure our windmill instance.
Credentials management
A minor pain point for any research project I've worked on are the limited options for sharing credentials with external collaborators. Typically, we rely on oauth to grant access to collaborators however we have circumstances where we want to securely share credentials for services without oauth e.g. database credentials or API keys. Windmill has solved this problem for us by enabling us to store our resources within windmill while keeping them encrypted. These resources are available directly to all the scripts within windmill and also to any other script running outside windmill such as the local environment.
Windmill provides tools for local development which can be used to test code locally before pushing it to your windmill instance but additionally, it also allows you to access the resources or credentials from your windmill instance. Resources can be fetched with a simple HTTP GET request using your API token in the header or directly within your local script.
pacs_credentials = wmill.get_resource("f/dicoms/trinidad_pacs")
db_credentials = json.loads(wmill.get_variable("f/kobo/vultr_db"))
curl -s -H "Authorization: Bearer $WM_TOKEN" \
"$BASE_INTERNAL_URL/api/w/$WM_WORKSPACE/resources/get_value_interpolated/f/dicoms/trinidad_pacs" | jq
Conclusion
Windmill provided a great gradual progression without requiring significant resources or vendor lock-in. While we haven't fully leveraged windmill's features, it has already been immensely helpful in saving us time and resources. Windmill has built-in support to use postgres transactions (INSERT, DELETE, UPDATE statements) to trigger jobs which we'd like to eventually shift to instead of the current method of using timed executions.
We're exploring Windmill's ML integrations, imagining systems that don't just move data but interpret it and automatically flag quality issues or generate preliminary analyses. As we continue to refine our workflows, we look forward to discovering other similar tools to improve our efficiencies!